-Search query
-Search result
Showing 1 - 50 of 686 items for (author: li & yc)

EMDB-41373: 
E. coli MraY mutant-T23P
Method: single particle / : Orta AK, Li YE, Clemons WM

PDB-8tlu: 
E. coli MraY mutant-T23P
Method: single particle / : Orta AK, Li YE, Clemons WM
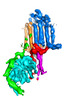
EMDB-40572: 
Human adenylyl Cyclase 5 in complex with Gbg
Method: single particle / : Yen YC, Tesmer JJG

EMDB-40573: 
Dimeric form of human adenylyl cyclase 5
Method: single particle / : Yen YC, Tesmer JJG

EMDB-40650: 
Phosphoinositide phosphate 3 kinase gamma
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40651: 
Phosphoinositide phosphate 3 kinase gamma bound with ATP
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40652: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40653: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and Gbetagamma
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40654: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and two Gbetagamma subunits in State 1
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40655: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and two Gbetagamma subunits in State 2
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-43658: 
SARS-CoV-2 S (C.37 Lambda variant) plus S309, S2L20, and S2X303 Fabs
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-43659: 
SARS-CoV-2 S NTD (C.37 Lambda variant) plus S2L20 and S2X303 Fabs, local refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-43660: 
SARS-CoV-2 S RBD (C.37 Lambda variant) plus S309 Fab, local refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-8vye: 
SARS-CoV-2 S (C.37 Lambda variant) plus S309, S2L20, and S2X303 Fabs
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-8vyf: 
SARS-CoV-2 S NTD (C.37 Lambda variant) plus S2L20 and S2X303 Fabs, local refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-8vyg: 
SARS-CoV-2 S RBD (C.37 Lambda variant) plus S309 Fab, local refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-36138: 
CrtSPARTA Octamer bound with guide-target
Method: single particle / : Guo LJ, Huang PP, Li ZX, Xiao YB, Chen MR

EMDB-36059: 
Short ago complexed with TIR-APAZ
Method: single particle / : Guo LJ, Huang PP, Li ZX, Xiao YB, Chen MR

EMDB-36070: 
SPARTA monomer bound with guide-target, state 2
Method: single particle / : Li ZX, Guo LJ, Huang PP, Xiao YB, Chen MR

EMDB-36095: 
CrtSPARTA hetero-dimer bound with guide-target, state 1
Method: single particle / : Li ZX, Guo LJ, Huang PP, Xiao YB, Chen MR

EMDB-36114: 
SPARTA dimer bound with guide-target
Method: single particle / : Li ZX, Guo LJ, Huang PP, Xiao YB, Chen MR

EMDB-34314: 
SARS-CoV-2 RNA E-RTC complex with RMP-nsp9 and GMPPNP
Method: single particle / : Yan LM, Huang YC, Ge J, Liu ZY, Gao Y, Rao ZH, Lou ZY

EMDB-16626: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium, 3'-phosphoadenosine-5'-phosphosulfate, and nanobody nAb7 (original map)
Method: single particle / : Mycroft-West CJ, Wu L

EMDB-16627: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium, 3'-phosphoadenosine-5'-phosphosulfate, and nanobody nAb7 (locally refined map of N-terminal and deacetylase domains)
Method: single particle / : Mycroft-West CJ, Wu L

EMDB-16629: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium, 3'-phosphoadenosine-5'-phosphosulfate, and nanobody nAb7 (locally refined map of deacetylase and sulfotransferase domains)
Method: single particle / : Mycroft-West CJ, Wu L

EMDB-16661: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium, 3'-phosphoadenosine-5'-phosphosulfate, and nanobody nAb13 (original map)
Method: single particle / : Mycroft-West CJ, Wu L

EMDB-16662: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium, 3'-phosphoadenosine-5'-phosphosulfate, and nanobody nAb13 (locally refined map of N-terminal and deacetylase domains)
Method: single particle / : Mycroft-West CJ, Wu L

EMDB-16663: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium, 3'-phosphoadenosine-5'-phosphosulfate, and nanobody nAb13 (locally refined map of deacetylase and sulfotransferase domains)
Method: single particle / : Mycroft-West CJ, Wu L

EMDB-16664: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium, 3'-phosphoadenosine-5'-phosphosulfate and nanobody nAb13 (composite map and model).
Method: single particle / : Mycroft-West CJ, Wu L

PDB-8chs: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium, 3'-phosphoadenosine-5'-phosphosulfate and nanobody nAb13 (composite map and model).
Method: single particle / : Mycroft-West CJ, Wu L

EMDB-16564: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium and 3'-phosphoadenosine-5'-phosphosulfate
Method: single particle / : Mycroft-West CJ, Wu L

EMDB-16565: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium, 3'-phosphoadenosine-5'-phosphosulfate, and nanobody nAb7 (composite map and model)
Method: single particle / : Mycroft-West CJ, Wu L

PDB-8ccy: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium and 3'-phosphoadenosine-5'-phosphosulfate
Method: single particle / : Mycroft-West CJ, Wu L

PDB-8cd0: 
Human heparan sulfate N-deacetylase-N-sulfotransferase 1 in complex with calcium, 3'-phosphoadenosine-5'-phosphosulfate, and nanobody nAb7 (composite map and model)
Method: single particle / : Mycroft-West CJ, Wu L
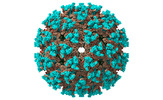
EMDB-15898: 
Recombinant Mayaro virus-like particle
Method: single particle / : Kim YC, Song X, Huiskonen JT

EMDB-34914: 
CXCR3-DNGi complex activated by CXCL11
Method: single particle / : Jiao HZ, Hu HL

EMDB-34915: 
CXCR3-DNGi complex activated by PS372424
Method: single particle / : Jiao HZ, Hu HL

EMDB-34916: 
CXCR3-DNGi complex activated by VUF11222
Method: single particle / : Jiao HZ, Hu HL

EMDB-34917: 
Structure of CXCR3 complexed with antagonist SCH546738
Method: single particle / : Jiao HZ, Hu HL

EMDB-36841: 
Structure of CXCR3 complexed with antagonist AMG487
Method: single particle / : Jiao HZ, Hu HL

EMDB-36842: 
CXCR3-DNGi complex activated by CXCL10
Method: single particle / : Jiao HZ, Hu HL
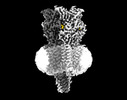
EMDB-16555: 
Mouse serotonin 5-HT3A receptor in complex with PZ-1922
Method: single particle / : Lopez-Sanchez U, Nury H
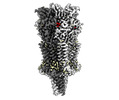
EMDB-16557: 
Mouse serotonin 5-HT3A receptor in complex with PZ-1939
Method: single particle / : Lopez-Sanchez U, Nury H

PDB-8cc6: 
Mouse serotonin 5-HT3A receptor in complex with PZ-1922
Method: single particle / : Lopez-Sanchez U, Nury H

PDB-8cc7: 
Mouse serotonin 5-HT3A receptor in complex with PZ-1939
Method: single particle / : Lopez-Sanchez U, Nury H

EMDB-40933: 
Structure of mouse Myomaker bound to Fab18G7 in detergent
Method: single particle / : Long T, Li X

EMDB-40934: 
Structure of mouse Myomaker bound to Fab18G7 in nanodiscs
Method: single particle / : Long T, Li X

EMDB-40935: 
Structure of Ciona Myomaker bound to Fab1A1
Method: single particle / : Long T, Li X

EMDB-40936: 
Structure of mouse Myomaker mutant-R107A bound to Fab18G7
Method: single particle / : Long T, Li X

EMDB-40937: 
Structure of mouse Myomaker mutant-Y118A bound to Fab18G7
Method: single particle / : Long T, Li X
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
